Jed Quiaoit
Samantha Himegarner
Jed Quiaoit
Samantha Himegarner
Skills you’ll gain in this topic:
- Explain the steps of transcription, from initiation to RNA synthesis.
- Describe RNA processing events, including splicing and polyadenylation.
- Relate mRNA processing to protein diversity and gene regulation.
- Predict effects of transcription mutations on protein synthesis.
- Analyze the role of RNA processing in gene expression.
Meet the RNA: The Transcription Process
Going back to the big idea relationship between structure and function, the sequence of the RNA bases, together with the structure of the RNA molecule, determines RNA function!
mRNA: Messenger RNA
mRNA molecules, also known as messenger RNA, are transcribed from DNA and carry genetic information from the DNA to the ribosome, where the information is translated into a primary peptide sequence. 💬
The sequence of bases in the mRNA molecule codes for the specific amino acid sequence of a protein. The structure of the mRNA molecule, including its secondary and tertiary structure, also plays a role in determining its function.
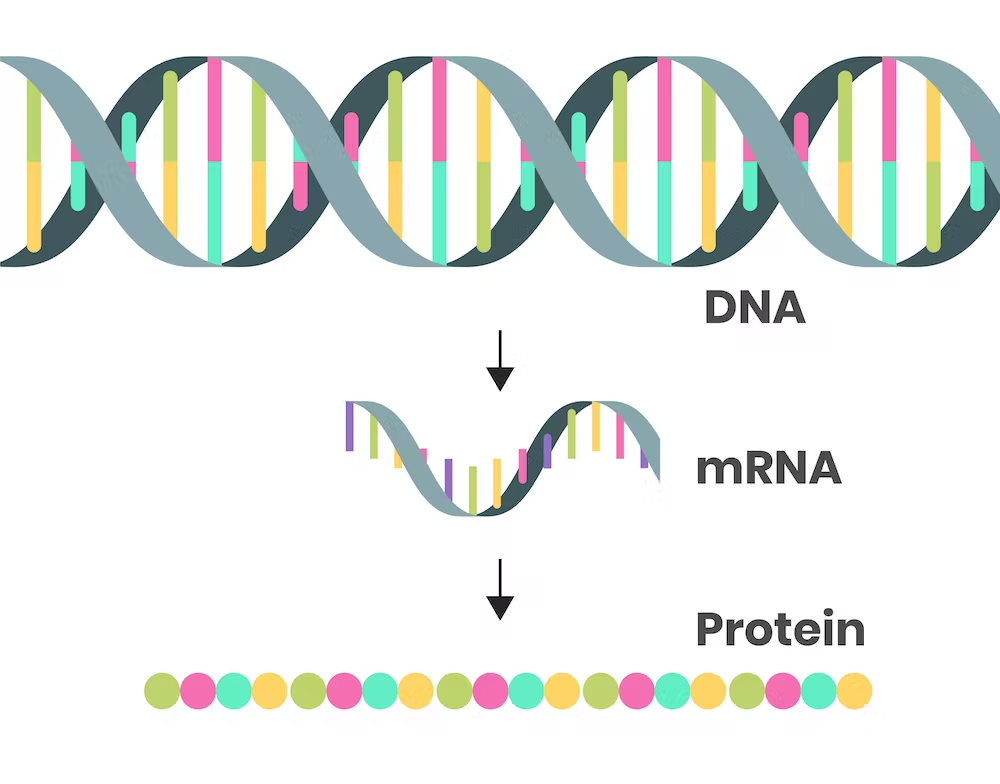
Source: The Conversation
tRNA: Transfer RNA
tRNA molecules, also known as transfer RNA, are small RNA molecules that function in protein synthesis. They have specific binding sites for specific amino acids, as well as an anticodon sequence that base pairs with the codon sequence of the mRNA.
During translation, tRNA molecules are recruited to the ribosome, where they bring the correct amino acid to be incorporated into the growing peptide chain based on the mRNA sequence. The structure of tRNA molecules also plays a role in their function, including the L-shaped conformation that allows them to bind both amino acids and the ribosome. 🚜
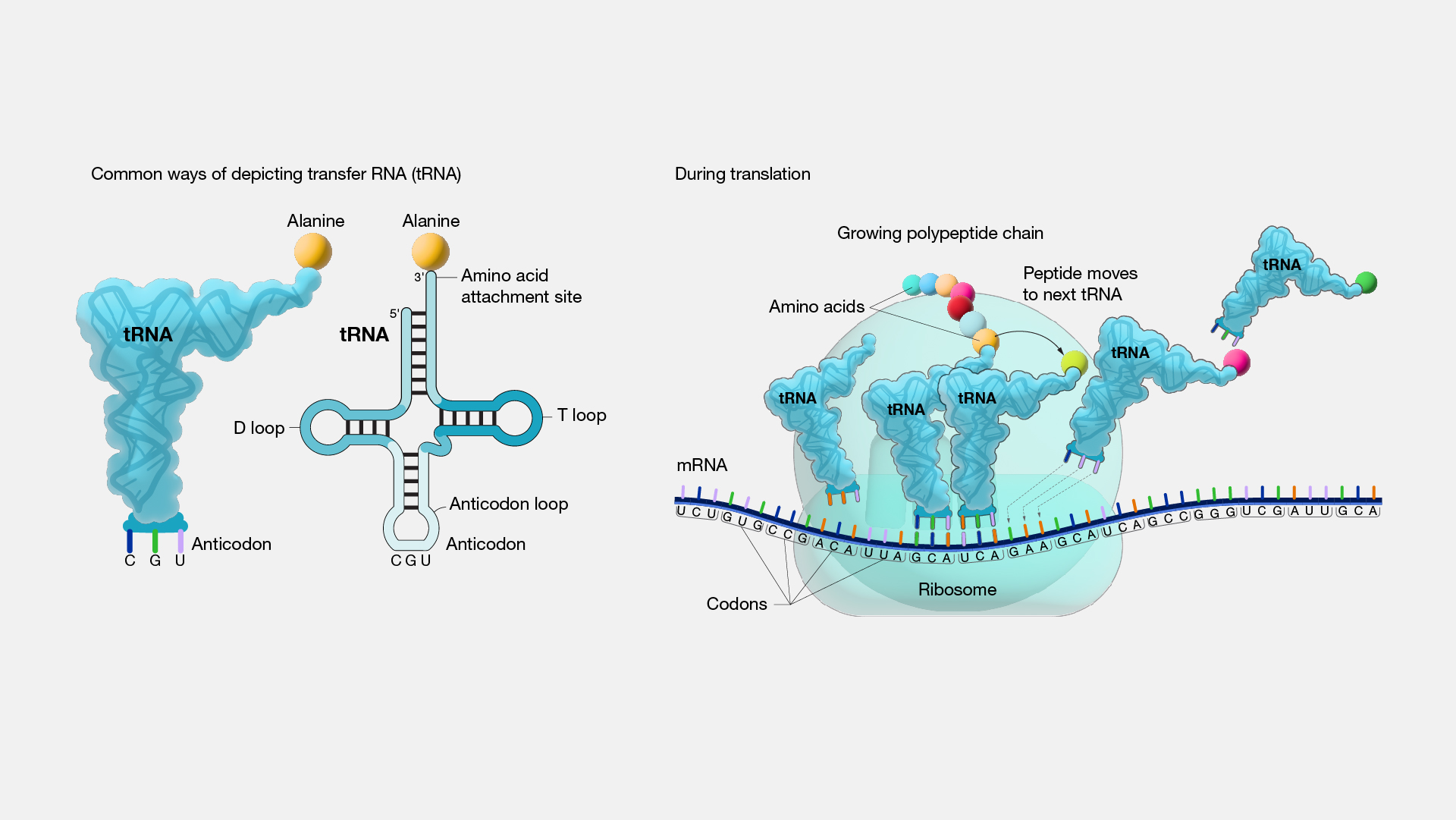
Source: National Human Genome Research Institute
rRNA: Ribosomal RNA
rRNA molecules, also known as ribosomal RNA, are the most abundant type of RNA in cells and are the functional building blocks of ribosomes. Ribosomes are the cellular structures that carry out protein synthesis, and rRNA molecules form the structural backbone of the ribosome.
They also interact with the mRNA and tRNA molecules during the translation process, helping to position the tRNA molecules and catalyze the peptide bond formation. The sequence and structure of rRNA molecules are also important for their function, as mutations in these regions can affect the efficiency and accuracy of protein synthesis. ⚙️
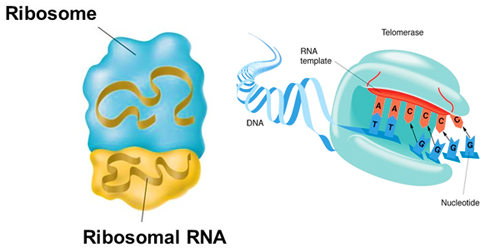
Source: Miami Bio
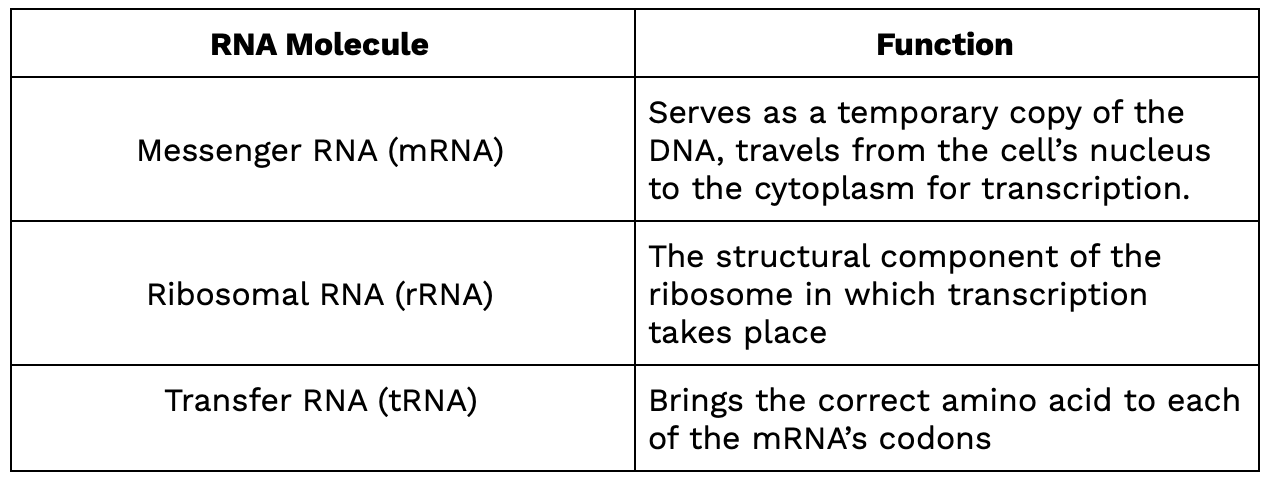
To summarize the three types of RNA:
The Central Dogma: DNA to RNA to Protein
Genetic information flows from DNA to protein through a process known as the central dogma of molecular biology. This process begins with the transcription of DNA into RNA, specifically messenger RNA (mRNA) molecules. During transcription, a specific portion of the DNA double helix, known as a gene, is copied into a complementary RNA molecule. 🐕
This process is catalyzed by an enzyme called RNA polymerase, which reads the DNA sequence and synthesizes a complementary RNA strand. The sequence of nucleotides in the DNA is used as a template to determine the sequence of bases in the mRNA molecule.
The sequence of bases in the mRNA molecule, known as the codon, carries the genetic information that codes for the specific amino acid sequence of a protein. This genetic information is translated into a protein through the process of translation. Translation, which we'll explore further in the next section, occurs on the ribosome, a large complex of RNA and protein molecules.
This process of transcription and translation results in the formation of a protein, with a specific sequence of amino acids determined by the sequence of nucleotides in the DNA. This protein can then perform a wide range of functions, including catalyzing metabolic reactions, replicating DNA, responding to stimuli, and transporting molecules across cell membranes.
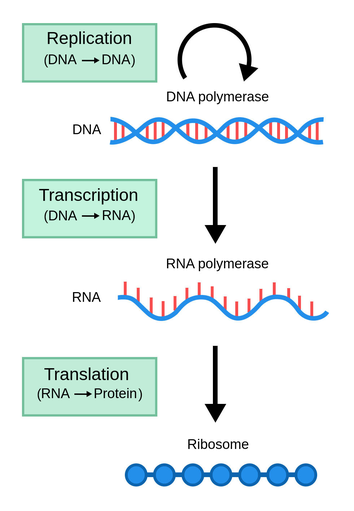
Source: Labster Theory
RNA Polymerase, the Ace of Transcription
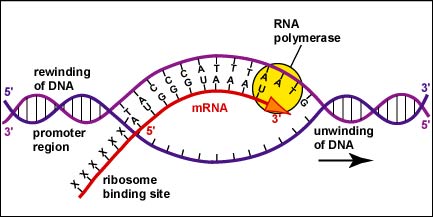
Again, transcription is the process by which a specific portion of DNA, known as a gene, is copied into a complementary RNA molecule. This process is catalyzed by an enzyme called RNA polymerase, which reads the DNA sequence and synthesizes a complementary RNA strand. 🪡
Source: Biology LibreTexts
The RNA polymerase binds to a specific region of the DNA called the promoter, which signals the start of the gene. The enzyme then moves along the template strand of DNA, reading the sequence of nucleotides and adding complementary nucleotides to the growing RNA strand. The process of transcription results in the formation of a primary RNA transcript, which can then undergo further processing to generate a mature RNA molecule.
During transcription, the RNA polymerase uses a single strand of DNA, known as the template strand, as a guide to determine the sequence of bases in the newly formed RNA molecule. This process is known as template-directed synthesis, and it results in the formation of an RNA molecule that is complementary to the template strand of DNA. The other strand of DNA, known as the coding strand, has the same sequence as the RNA, except that thymine is replaced by uracil.
Template Strand
The DNA strand that serves as the template strand during transcription is also referred to as the noncoding strand, minus strand, or antisense strand. This is because the sequence of the noncoding strand is not directly translated into a protein, unlike the coding strand which has the same sequence as the mRNA, except that thymine is replaced by uracil. The selection of which DNA strand serves as the template strand during transcription depends on the gene being transcribed.
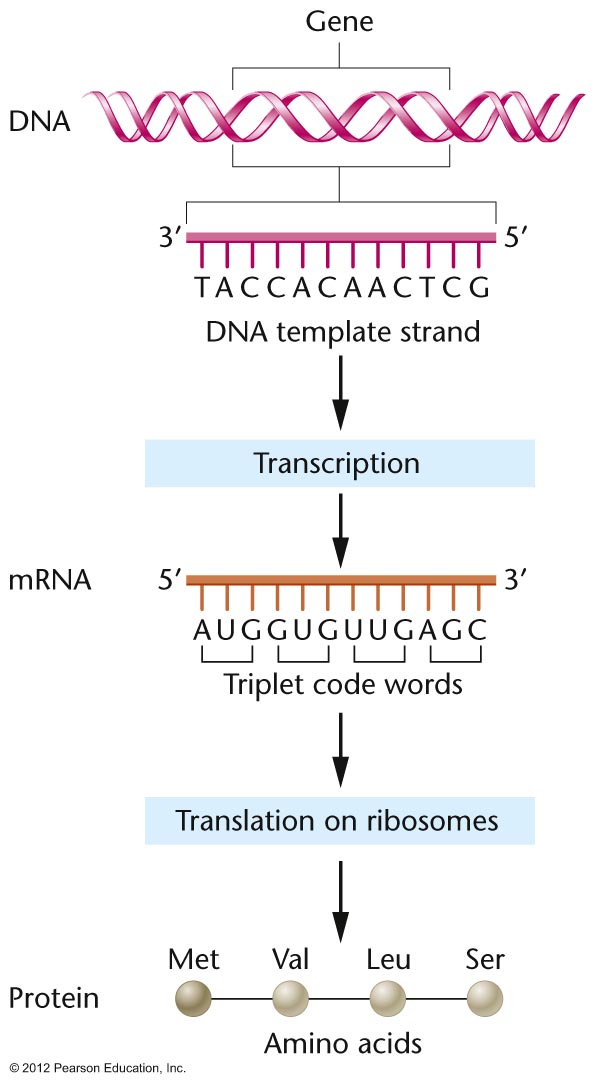
Source: Samford University
Not included in the AP Bio curriculum but interesting to compare and contrast: In prokaryotic cells, transcription usually occurs on the template strand that runs in the opposite direction as the coding strand, also known as the antisense strand. This is known as divergent transcription, where the coding and non-coding strands are oriented in opposite directions from one another.
In eukaryotic cells, which are cells with a defined nucleus, the majority of transcription occurs on the template strand that runs in the same direction as the coding strand, also known as the sense strand. This is known as convergent transcription, where the coding and non-coding strands are oriented in the same direction. However, there are also examples of divergent transcription in eukaryotic cells too.
The selection of the template strand can also be determined by certain regions of the DNA called regulatory elements, such as enhancers and promoters. These regions can bind to transcription factors, which are proteins that help to control the transcription process. The binding of these factors can determine which strand will be used as the template strand, and can also affect the rate and efficiency of transcription. 🚗💨
Processing the mRNA Transcript
In eukaryotic cells, the primary RNA transcript undergoes a series of enzyme-regulated modifications to generate a mature mRNA molecule that is ready for translation. These modifications include: 🔧
a. Addition of a poly-A tail: The addition of a string of adenine nucleotides, known as a poly-A tail, to the 3' end of the RNA transcript. This process is catalyzed by an enzyme called polyadenylate polymerase and it helps to protect the mRNA from degradation and also helps in the transport of mRNA out of the nucleus.
b. Addition of a GTP cap: The addition of a modified nucleotide, known as a GTP cap, to the 5' end of the RNA transcript. This process is catalyzed by an enzyme called guanylyltransferase (no need to remember for the AP exam). The cap helps to protect the mRNA from degradation, and also helps in the recognition of the mRNA by the translation machinery.
c. Excision of introns and splicing and retention of exons: The primary RNA transcript contains both coding regions (exons) and non-coding regions (introns) . The introns are removed by a process called RNA splicing. This process is catalyzed by a complex of enzymes called the spliceosome. The exons are then joined together to form a mature mRNA molecule that contains only the coding regions.
d. Excision of introns and splicing and retention of exons can generate different versions of the resulting mRNA molecule; this is known as alternative splicing. This occurs when different combinations of exons are joined together to form different mRNA molecules, each coding for a different protein or a different version of a protein. This process allows a single gene to produce multiple proteins and can increase the diversity of the protein products of a gene.
Alternative splicing is a major mechanism of gene regulation in eukaryotic cells and it plays a key role in the complexity and diversity of eukaryotic organisms.
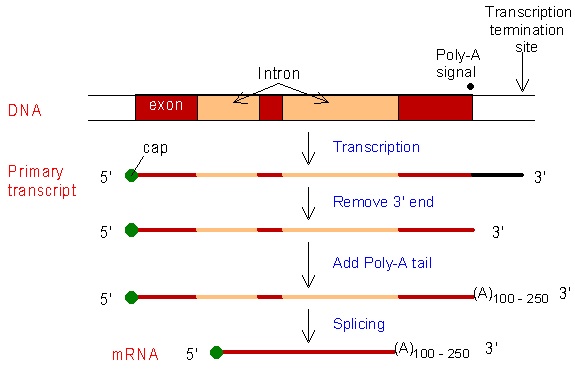
Source: Medbullets
** Check out the AP Bio Unit 6 Replays or watch the 2021 Unit 6 Cram**
<< Hide Menu
Jed Quiaoit
Samantha Himegarner
Jed Quiaoit
Samantha Himegarner
Skills you’ll gain in this topic:
- Explain the steps of transcription, from initiation to RNA synthesis.
- Describe RNA processing events, including splicing and polyadenylation.
- Relate mRNA processing to protein diversity and gene regulation.
- Predict effects of transcription mutations on protein synthesis.
- Analyze the role of RNA processing in gene expression.
Meet the RNA: The Transcription Process
Going back to the big idea relationship between structure and function, the sequence of the RNA bases, together with the structure of the RNA molecule, determines RNA function!
mRNA: Messenger RNA
mRNA molecules, also known as messenger RNA, are transcribed from DNA and carry genetic information from the DNA to the ribosome, where the information is translated into a primary peptide sequence. 💬
The sequence of bases in the mRNA molecule codes for the specific amino acid sequence of a protein. The structure of the mRNA molecule, including its secondary and tertiary structure, also plays a role in determining its function.

Source: The Conversation
tRNA: Transfer RNA
tRNA molecules, also known as transfer RNA, are small RNA molecules that function in protein synthesis. They have specific binding sites for specific amino acids, as well as an anticodon sequence that base pairs with the codon sequence of the mRNA.
During translation, tRNA molecules are recruited to the ribosome, where they bring the correct amino acid to be incorporated into the growing peptide chain based on the mRNA sequence. The structure of tRNA molecules also plays a role in their function, including the L-shaped conformation that allows them to bind both amino acids and the ribosome. 🚜

Source: National Human Genome Research Institute
rRNA: Ribosomal RNA
rRNA molecules, also known as ribosomal RNA, are the most abundant type of RNA in cells and are the functional building blocks of ribosomes. Ribosomes are the cellular structures that carry out protein synthesis, and rRNA molecules form the structural backbone of the ribosome.
They also interact with the mRNA and tRNA molecules during the translation process, helping to position the tRNA molecules and catalyze the peptide bond formation. The sequence and structure of rRNA molecules are also important for their function, as mutations in these regions can affect the efficiency and accuracy of protein synthesis. ⚙️

Source: Miami Bio
To summarize the three types of RNA:

The Central Dogma: DNA to RNA to Protein
Genetic information flows from DNA to protein through a process known as the central dogma of molecular biology. This process begins with the transcription of DNA into RNA, specifically messenger RNA (mRNA) molecules. During transcription, a specific portion of the DNA double helix, known as a gene, is copied into a complementary RNA molecule. 🐕
This process is catalyzed by an enzyme called RNA polymerase, which reads the DNA sequence and synthesizes a complementary RNA strand. The sequence of nucleotides in the DNA is used as a template to determine the sequence of bases in the mRNA molecule.
The sequence of bases in the mRNA molecule, known as the codon, carries the genetic information that codes for the specific amino acid sequence of a protein. This genetic information is translated into a protein through the process of translation. Translation, which we'll explore further in the next section, occurs on the ribosome, a large complex of RNA and protein molecules.
This process of transcription and translation results in the formation of a protein, with a specific sequence of amino acids determined by the sequence of nucleotides in the DNA. This protein can then perform a wide range of functions, including catalyzing metabolic reactions, replicating DNA, responding to stimuli, and transporting molecules across cell membranes.

Source: Labster Theory
RNA Polymerase, the Ace of Transcription
Again, transcription is the process by which a specific portion of DNA, known as a gene, is copied into a complementary RNA molecule. This process is catalyzed by an enzyme called RNA polymerase, which reads the DNA sequence and synthesizes a complementary RNA strand. 🪡

Source: Biology LibreTexts
The RNA polymerase binds to a specific region of the DNA called the promoter, which signals the start of the gene. The enzyme then moves along the template strand of DNA, reading the sequence of nucleotides and adding complementary nucleotides to the growing RNA strand. The process of transcription results in the formation of a primary RNA transcript, which can then undergo further processing to generate a mature RNA molecule.
During transcription, the RNA polymerase uses a single strand of DNA, known as the template strand, as a guide to determine the sequence of bases in the newly formed RNA molecule. This process is known as template-directed synthesis, and it results in the formation of an RNA molecule that is complementary to the template strand of DNA. The other strand of DNA, known as the coding strand, has the same sequence as the RNA, except that thymine is replaced by uracil.
Template Strand
The DNA strand that serves as the template strand during transcription is also referred to as the noncoding strand, minus strand, or antisense strand. This is because the sequence of the noncoding strand is not directly translated into a protein, unlike the coding strand which has the same sequence as the mRNA, except that thymine is replaced by uracil. The selection of which DNA strand serves as the template strand during transcription depends on the gene being transcribed.

Source: Samford University
Not included in the AP Bio curriculum but interesting to compare and contrast: In prokaryotic cells, transcription usually occurs on the template strand that runs in the opposite direction as the coding strand, also known as the antisense strand. This is known as divergent transcription, where the coding and non-coding strands are oriented in opposite directions from one another.
In eukaryotic cells, which are cells with a defined nucleus, the majority of transcription occurs on the template strand that runs in the same direction as the coding strand, also known as the sense strand. This is known as convergent transcription, where the coding and non-coding strands are oriented in the same direction. However, there are also examples of divergent transcription in eukaryotic cells too.
The selection of the template strand can also be determined by certain regions of the DNA called regulatory elements, such as enhancers and promoters. These regions can bind to transcription factors, which are proteins that help to control the transcription process. The binding of these factors can determine which strand will be used as the template strand, and can also affect the rate and efficiency of transcription. 🚗💨
Processing the mRNA Transcript
In eukaryotic cells, the primary RNA transcript undergoes a series of enzyme-regulated modifications to generate a mature mRNA molecule that is ready for translation. These modifications include: 🔧
a. Addition of a poly-A tail: The addition of a string of adenine nucleotides, known as a poly-A tail, to the 3' end of the RNA transcript. This process is catalyzed by an enzyme called polyadenylate polymerase and it helps to protect the mRNA from degradation and also helps in the transport of mRNA out of the nucleus.
b. Addition of a GTP cap: The addition of a modified nucleotide, known as a GTP cap, to the 5' end of the RNA transcript. This process is catalyzed by an enzyme called guanylyltransferase (no need to remember for the AP exam). The cap helps to protect the mRNA from degradation, and also helps in the recognition of the mRNA by the translation machinery.
c. Excision of introns and splicing and retention of exons: The primary RNA transcript contains both coding regions (exons) and non-coding regions (introns) . The introns are removed by a process called RNA splicing. This process is catalyzed by a complex of enzymes called the spliceosome. The exons are then joined together to form a mature mRNA molecule that contains only the coding regions.
d. Excision of introns and splicing and retention of exons can generate different versions of the resulting mRNA molecule; this is known as alternative splicing. This occurs when different combinations of exons are joined together to form different mRNA molecules, each coding for a different protein or a different version of a protein. This process allows a single gene to produce multiple proteins and can increase the diversity of the protein products of a gene.
Alternative splicing is a major mechanism of gene regulation in eukaryotic cells and it plays a key role in the complexity and diversity of eukaryotic organisms.

Source: Medbullets
** Check out the AP Bio Unit 6 Replays or watch the 2021 Unit 6 Cram**

© 2024 Fiveable Inc. All rights reserved.